HistoPlexer predicts protein maps from H&E images
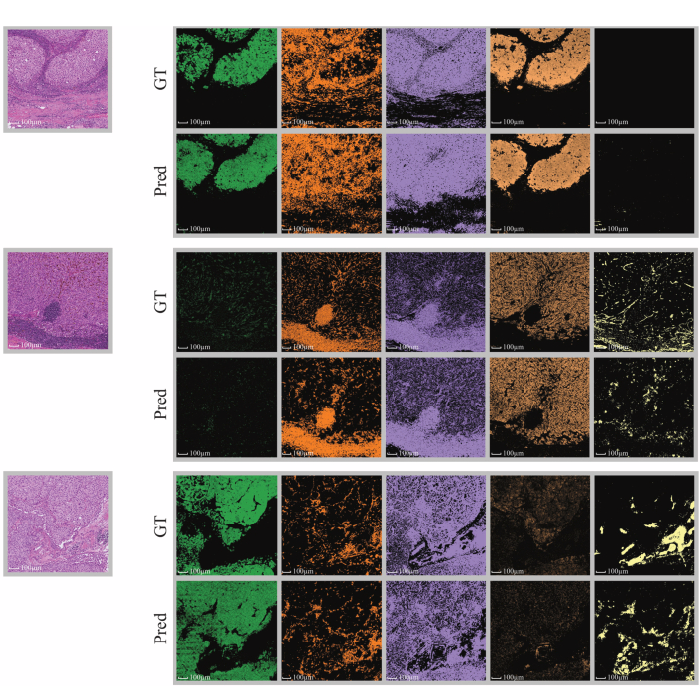
Nature Machine Intelligence has published HistoPlexer, a conditional GAN that produces multiplexed protein maps from standard H&E, outperforming baseline methods and preserving marker co localization across metastatic melanoma and public datasets. Expert readers frequently mistook generated images for real, underscoring realism.
The model scales to whole slide images, supports immune phenotyping based on predicted CD8+ T cell distributions, and generalizes to TCGA SKCM. Adding HistoPlexer derived features boosted survival prediction C index by 3.18% and increased weighted F1 for immune subtype classification by 17.02%.
DQBM’s Bodenmiller Lab contributed data and multiplex imaging expertise through the Tumor Profiler Consortium, with Bernd Bodenmiller as co author and contributor of tumour material.
DOI: https://doi.org/10.1038/s42256-025-01074-y