Navigation auf uzh.ch
Navigation auf uzh.ch
See Mathis et al., Nature Biotechnology 2024
The success of prime editing depends on the prime editing guide RNA (pegRNA) design and target locus. Here, we developed machine learning models that reliably predict prime editing efficiency. PRIDICT2.0 assesses the performance of pegRNAs for all edit types up to 15 bp in length in mismatch repair-deficient and mismatch repair-proficient cell lines and in vivo in primary cells. With ePRIDICT, we further developed a model that quantifies how local chromatin environments impact prime editing rates.
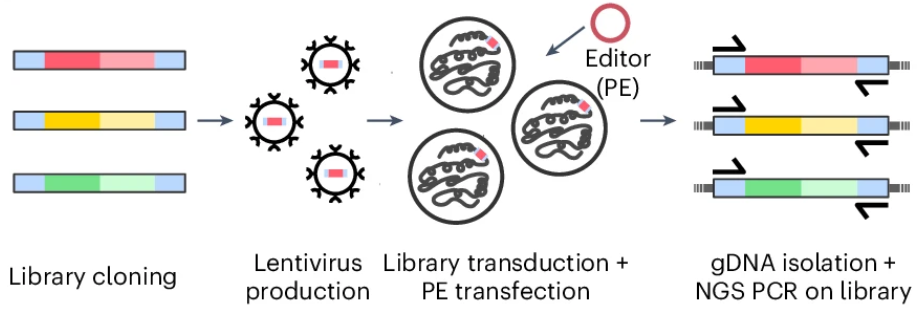
Schematic overview of the screen with the target-matched pegRNA library Library-Diverse.