News
News list
-

Support of applicants for the SNSF Professorial Fellowship / SNSF Starting Grant in the field of "Quantitative Biomedicine"
The Department of Quantitative Biomedicine (DQBM) of the University of Zurich announces opportunities for applicants who seek to establish their own research group by obtaining independent third-party funding (SNSF Professorial Fellowship / SNSF Starting Grant).
Find all information here.
-
The Krauthammer Group Develops Explainable Deep Learning Model for Predicting Disease Activity in Joint Disorders
See Trottet et al., PLOS Digital Health 2024
A new study by the Krauthammer group presents DAS-Net, an explainable deep learning model for predicting disease activity in chronic inflammatory joint diseases. This model utilizes patient data to make predictions and identifies similar patients based on disease progression. The approach outperforms traditional models and enhances understanding through feature attribution, providing insights into key patient characteristics impacting disease.
-
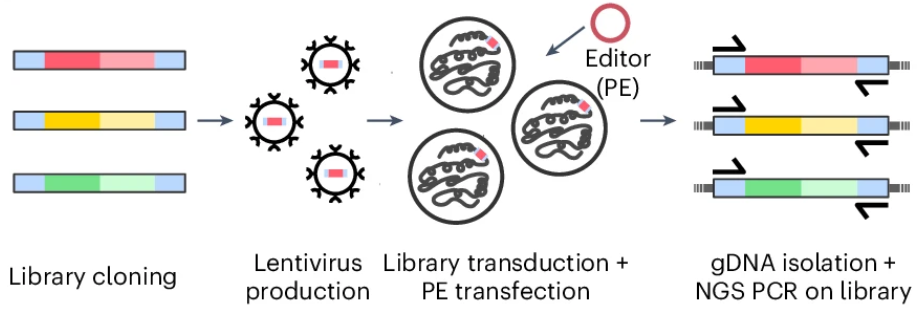
The Krauthammer group and collaborators develop advanced AI model to predict disease risk from multimodal data
See Mathis et al., Nature Biotechnology 2024
The success of prime editing depends on the prime editing guide RNA (pegRNA) design and target locus. Here, we developed machine learning models that reliably predict prime editing efficiency. PRIDICT2.0 assesses the performance of pegRNAs for all edit types up to 15 bp in length in mismatch repair-deficient and mismatch repair-proficient cell lines and in vivo in primary cells. With ePRIDICT, we further developed a model that quantifies how local chrom
-
In collaboration, the Joller group and the Krauthammer group reveal the regulatory role of tissue-specific Tregs in autoimmune diseases
See Rakebrandt et al., Proc Natl Acad Sci U S A. 2024
The Krauthammer group introduces Fragmentstein, a command-line tool designed to convert non-sensitive cfDNA fragment data into BAM files suitable for standard bioinformatics analyses. This innovative approach facilitates data sharing without compromising sensitive genomic information, enabling analyses like CNV, nucleosome occupancy, and fragment length studies while ensuring data privacy.
-

Bernd Bodenmiller on strategy& Insider Podcast on Harnessing AI for next generation cancer diagnostics
DQBM's Prof. Dr. Bernd Bodenmiller appears on the latest issue of the strategy& insider podcast, talking about Harnessing AI for next generation cancer diagnostics. In this episode, Bernd Bodenmiller and Andreas Wicki share their vision to transform cancer diagnosis, treatment, and management through cutting-edge tumor profiling techniques and exploratory research. Tune in and find out about recent developments in cancer research, the power of data and technology – and how AI can revolutionize precision oncology.
Tune in now on Apple Podcasts, Spotify, or Podigee.
-
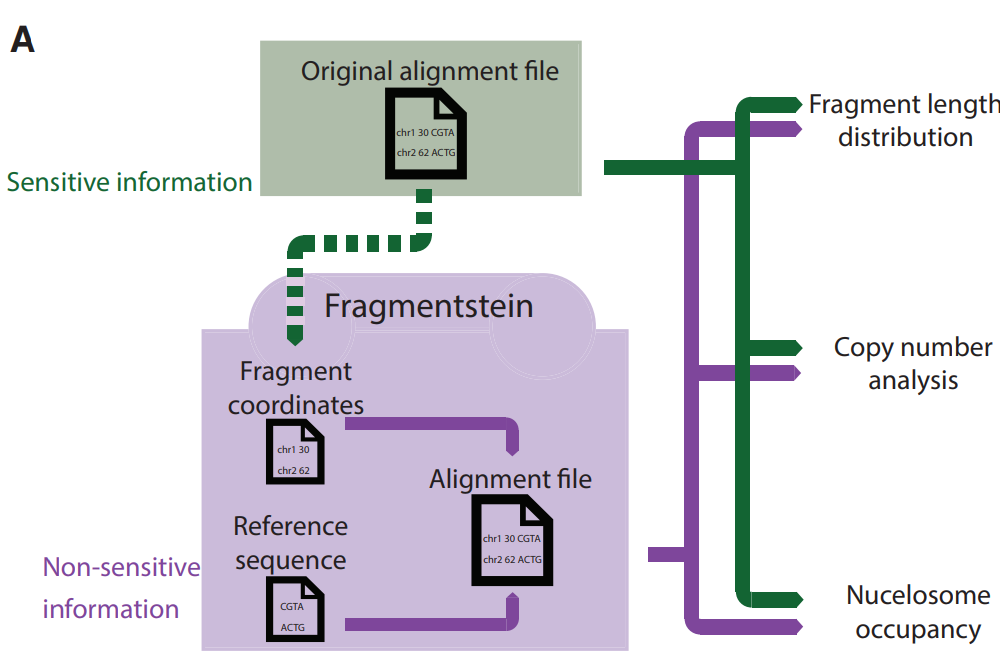
The Krauthammer group develops Fragmentstein: A Novel Tool for cfDNA Fragment Analysis
See Balázs et al., Bioinformatics 2024
The Krauthammer group introduces Fragmentstein, a command-line tool designed to convert non-sensitive cfDNA fragment data into BAM files suitable for standard bioinformatics analyses. This innovative approach facilitates data sharing without compromising sensitive genomic information, enabling analyses like CNV, nucleosome occupancy, and fragment length studies while ensuring data privacy.
-

The Polymenidou group and collaborators detect TDP-43 seeding activity in the olfactory mucosa of FTD patients
See Fontana and Bongianni et al., Alzheimer's and Dementia
In this work, the Polymenidou group and collaborators assessed TAR DNA-binding protein 43 (TDP-43) seeding activity and aggregates detection in olfactory mucosa of patients with frontotemporal lobar degeneration with TDP-43-immunoreactive pathology (FTLD-TDP) by TDP-43 seeding amplification assay (TDP43-SAA) and immunocytochemical analysis. They demonstrate that TDP-43 aggregates can be detectable in olfactory mucosa, suggesting that their TDP43-Seeding Amplification Assay might be useful for identifying and monitoring FTLD-TDP in living patients.
-

The Bodenmiller group develops an end-to-end multiplexed image processing and analysis workflow
See Windhager et al., Nature Protocols
In this work, the Bodenmiller group presents an end-to-end workflow for multiplexed tissue image processing and analysis that integrates previously developed computational tools to enable image segmentation, feature extraction and spatially resolved single-cell analysis in a user-friendly and customizable way. This protocol can be implemented by researchers with basic bioinformatics training, and the analysis of the provided dataset can be completed within 5–6 h.
An extended version is available at https://bodenmillergroup.github.io/IMCDataAnalysis/. -

The Menze group develops diffusion-based hierarchical multi-label object detection to analyze panoramic dental X-Rays
See Hamamci et al., MICCAI 2023
Numerous Machine Learning models for the interpretation of panoramic dental X-rays have been developed, yet none of them offers an end-to-end solution that identifies problematic teeth with dental enumeration and associated diagnoses at the same time. In this work, the Menze group structure three distinct types of annotated data hierarchically following the FDI system. To learn from all three hierarchies jointly, a novel diffusion-based hierarchical multi-label object detection framework is introduced by adapting a diffusion-based method that formulates object detection as a denoising diffusion process from noisy boxes to object boxes. Experimental results show that this novel method significantly outperforms state-of-the-art object detection methods.
The code and the datasets are available at https://github.com/ibrahimethemhamamci/HierarchicalDet. -

The Menze group develops a self-pruning graph neural network for predicting inflammatory disease activity in Multiple Sclerosis from Brain MR Images
See Prabhakar et al., MICCAI 2023
Multiple Sclerosis (MS) is a severe neurological disease characterized by inflammatory lesions in the central nervous system. Predicting inflammatory disease activity is crucial for disease assessment and treatment. In this work, the Menze group presents the first attempt to utilize graph neural networks (GNN) to aggregate MS biomarkers for a novel global representation. A two-stage MS inflammatory disease activity prediction approach is proposed that detects lesions using a 3D segmentation network and extracts their image features using a self-supervised algorithm. The detected lesions are used to build a patient graph, where the lesions act as nodes that are connected based on spatial proximity and the inflammatory disease activity prediction is formulated as a graph classification task. A self-pruning strategy auto-selects the most critical lesions for prediction. The proposed method outperforms the existing baseline by a large margin and offers inherent explainability by assigning an importance score to each lesion for the overall prediction.
Code is available at https://github.com/chinmay5/ms_ida.git. -

DQBM and the Institute for Implementation Science in Healthcare (IfIS) launch BME337: "Introduction to Digital Health"
DQBM and the Institute for Implementation Science in Healthcare (IfIS) launch BME337: "Introduction to Digital Health" this Autumn semester. This course is open to MNF and Mef students alike.
Link to BME337: Introduction to Digital Health in the course catalogue
This course will provide an overview of tools, methods and applications driving digital health innovations. ln the course, students will gain first insights into the topic by UZH digital health experts presenting their latest research in clinical data science, imaging, mobile health, digital therapeutics, augmented reality and related domains. The course is the first one of five modules on 'Digital Heath', which aims at providing students with the fundamental knowledge about using data and computation in clinical research and digital health interventions. To prepare the students for the following modules, this introductory course will provide the necessary understanding of the key concepts in digital health at the intersection of (bio- and behavioural) medicine, data science, bioinformatics, and information systems.Topics that will be introduced in this course include:
- General overview on how data, digital tools and Al are used in healthcare;
- Clinical data science and translational informatics;
- Biomedical lmage Analysis and Machine Learning;
- Digital Tools for Medical Knowledge and Decision Support;
- Mobile health data streams for real world evidence generation and public health;
- Digital therapeutics and health interventions for prevention, management, and treatment of diseases.
-

The Bodenmiller group develops DNA-barcoded signal amplification for imaging mass cytometry
DNA-barcoded signal amplification for imaging mass cytometry enables sensitive and highly multiplexed tissue imaging
See Hosogane et al., Nature Methods
In this work, the Bodenmiller group extends Imaging mass cytometry (IMC) to low-abundance markers through incorporation of the DNA-based signal amplification by exchange reaction, immuno-SABER. Using this novel method, the tumor immune microenvironment in human melanoma was analyzed by simultaneous imaging of 18 markers with immuno-SABER and 20 markers without amplification. SABER-IMC enabled the identification of immune cell phenotypic markers that are not detectable with IMC alone.
-

The Bodenmiller group develops multielement Z-tag imaging by X-ray fluorescence microscopy
Multielement Z-tag imaging by X-ray fluorescence microscopy for next-generation multiplex imaging
See Strotton et al., Nature Methods
Whole-organism to atomic-level imaging is possible with tissue-penetrant, picometer-wavelength X-rays. To enable highly multiplexed X-ray imaging, the Bodenmiller group developed multielement Z-tag X-ray fluorescence (MEZ-XRF) that can operate at kHz speeds when combined with signal amplification by exchange reaction (SABER)-amplified Z-tag reagents. Parallel imaging of 20 Z-tag or SABER Z-tag reagents at subcellular resolution was demonstrated in cell lines and multiple human tissues. The unique multiscale, nondestructive nature of MEZ-XRF, combined with SABER Z-tags for high sensitivity or enhanced speed, enables highly multiplexed bioimaging across biological scales.
-

The Kümmerli group explores bacterial strain-specific resistance evolution
Rapid and strain-specific resistance evolution of Staphylococcus aureus against inhibitory molecules secreted by Pseudomonas aeruginosa
See Niggli et al., mBio
Polymicrobial infections are widespread and pathogenoc interaction potentially trigger evolutionary dynamics with pathogens adapting to each other. In this work, the Kümmerli group explored the potential of Staphylococcus aureus to adapt to its competitor Pseudomonas aeruginosa. Experimental evolution experiments with three different S. aureus strains showed that S. aureus rapidly becomes resistant to inhibitory compounds secreted by P. aeruginosa. Three main factors contribute to strain-specific resistance evolution: (i) overproduction of a molecule that protects against oxidative stress; (ii) the formation of small colony variants that also protect against oxidative stress; and (iii) changes in membrane transporters that may reduce toxin uptake. Taken together, this work shows that species interactions can change over time, potentially favoring species coexistence, which in turn could influence disease progression and treatment options. -

The Menze group develops a method to generate accurate synthetic CT images from MRI data
Region of interest focused MRI to synthetic CT translation using regression and segmentation multi-task network
See Kaushik et al., Phys Med BiolIn clinical workflow, replacing CT with MR image enhances workflow efficiency and reduces patient radiation. To eliminate CT from the workflow, the information provided by CT needs to be generated via an MR image. In this work, the Menze group proposes a machine learning method to generate accurate synthetic CT (sCT) from MRI with a quantitative accuracy suitable for RT dose planning application, setting the stage for a broader clinical evaluation of sCT based RT planning on different anatomical regions.
-

Michael Krauthammer takes over the DQBM Directorship from Bernd Bodenmiller per 01.08.2023
After 4,5 years, DQBM's Founding Director Prof. Dr. Bernd Bodenmiller has concluded his term as Head of Department, passing the gavel to Prof. Dr. Dr. Michael Krauthammer.
We thank Bernd for his service and congratulate Michael on his new role! -

The Kümmerli group shows that negative interactions and virulence differences drive polymicrobial infection dynamics
Negative interactions and virulence differences drive the dynamics in multispecies bacterial infections
See Schmitz et al., Proc Biol Sci.
Bacterial infections are often polymicrobial, leading to intricate pathogen-pathogen and pathogen-host interactions. Here, the Kümmerli group co-infected larvae of Galleria mellonella with one to four opportunistic human pathogens to show that host mortality is always determined by the most virulent bacterial species, regardless of the number of species and pathogen combinations injected. This work reveals positive associations between a pathogen's growth inside the host, its competitiveness towards other pathogens and its virulence, supporting the experimentally validated prediction that treatments against polymicrobial infections should first target the most virulent species to reduce host morbidity. -

The Bodenmiller group publishes a cancer-associated fibroblast classification scheme
Cancer-associated fibroblast classification in single-cell and spatial proteomics data
See Cords et al., Nature CommunicationsIn this work, the Bodenmiller group defined cancer-associated fibroblast (CAF) phenotypes by analysing a single-cell RNA sequencing (scRNA-seq) dataset of over 16,000 stromal cells from tumours of 14 breast cancer patients, based on which nine CAF phenotypes and one class of pericytes were defined and functionally annotated. Next, the classification system was validated in four additional cancer types and highly multiplexed imaging mass cytometry was used on matched breast cancer samples to confirm the defined CAF phenotypes at the protein level and to analyse their spatial distribution within tumours. This general CAF classification scheme will allow comparison of CAF phenotypes across studies, facilitate analysis of their functional roles, and potentially guide development of new treatment strategies in the future.
-

The Polymenidou group shows that loss of TDP-43 oligomerization or RNA binding elicits distinct protein aggregation patterns
Loss of TDP-43 oligomerization or RNA binding elicits distinct aggregation patterns
See Pérez-Berlanga et al., The EMBO Journal
In disease, the RNA-binding protein TAR DNA-binding protein 43 (TDP-43) is known to form cytoplasmic or intranuclear inclusions, but how TDP-43 transitions from physiological to pathological states remains poorly understood. In this work, the Polymenidou group unravels the origins of heterogeneous pathological species reminiscent of those occurring in TDP-43 proteinopathy patients. By using human neurons and cell lines with near-physiological expression levels, oligomerization and RNA binding are shown to govern TDP-43 stability, splicing functionality, LLPS, and subcellular localization. Monomeric TDP-43 is found to form inclusions in the cytoplasm, whereas its RNA binding-deficient counterpart aggregates in the nucleus. These differentially localized aggregates emerge via distinct pathways: LLPS-driven aggregation in the nucleus and aggresome-dependent inclusion formation in the cytoplasm. -
Read Michael Krauthammer's conversation with Schweizerische Ärztezeitung on AI in medicine (in German)
Wie wird die Künstliche Intelligenz (KI) die Medizin verändern? Ein Gespräch mit Medizininformatik-Professor Michael Krauthammer über KI, die Medizinprüfungen besteht und künftig vielleicht eigene Definitionen von Gesundheit und Krankheit entwickelt.
https://doi.org/10.4414/saez.2023.21842Read more about the new BioMedical Informatics Platform in UZH News: "Unlocking the Data Treasure Chest"
-

The Bodenmiller group identifies prognostic single-cell populations in breast cancer lymph node metastases
Multiplex imaging of breast cancer lymph node metastases identifies prognostic single-cell populations independent of clinical classifiers
In this work, the Bodenmiller group uses multiplex imaging mass cytometry to analyze breast cancer patients' single tumor cell phenotypes, both in primary tumors and in matched lymph node metastases. They report high phenotypic variability between these tissue sites and identify high- and low-risk disseminated cell phenotypes and prognostic markers.
See Fischer et al., Cell Reports Medicine -

The Bodenmiller lab and collaborators receive 2 mCHF for the CCCZ Lighthouse Project "IMMUNO-CAR ZURICH"
IMMUNO-CAR ZURICH (ZURICAR) aims to develop innovative platforms within the Comprehensive Cancer Center Zurich (CCCZ) for the effective, flexible, safe and cost-efficient production of Chimeric Antigen Receptor (CAR) immune cells, which will be manufactured at Wyss Zurich and subsequently used in Phase I clinical trials in patients with an urgent need for effective therapies. This highly translational, patient-centered approach is accompanied by state-of-the-art immune monitoring during therapy and research to improve the efficacy of CAR cells.
Read more about the IMMUNO-CAR ZURICH project on the CCCZ website -

Magdalini Polymenidou becomes Novartis Institutes for BioMedical Research (NIBR) Global Scholar
Congratulations to Prof. Dr. Magdalini Polymenidou, who was selected to become Novartis Institutes for BioMedical Research (NIBR) Global Scholar. The NIBR Global Scholars Program (NGSP) is a competitive program that seeks to advance science by supporting projects focused on novel science with the objective of being translated to drug discovery and/or clinical research. Magdalini Polymenidou has been selected for the NGSP 2022 and will receive up to 1 mil USD funding over three years along with scientific expertise from NIBR collaborators.
Please read the full UZH News article here.
Photo: Magdalini Polymenidou (by Pascal Halder www.naturphotos.ch)
-

The Menze group publishes a deep learning approach to predict collateral flow in stroke patients
A deep learning approach to predict collateral flow in stroke patients using radiomic features from perfusion images
In this work, the Menze group presents a multi-stage deep learning approach to predict collateral flow grading in stroke patients. Based on radiomic features extracted from MR perfusion data, a region of interest (RoI) detection task is formulated as a reinforcement learning problem and used to train a deep learning network to automatically detect the occluded region within the 3D MR perfusion volumes. Next, radiomic features are extracted from the obtained RoI through local image descriptors and denoising auto-encoders. Finally, a convolutional neural network and other machine learning classifiers are applied to the extracted radiomic features to automatically predict the collateral flow grading of the given patient volume as one of three severity classes. This automated deep learning approach is faster than visual inspection, eliminates grading bias and demonstrates a performance comparable to expert grading.
See Tetteh et al., Front Neurol
-

The Menze group publishes Focused Decoder: a novel Detection Transformer for 3D anatomical structure detection
Focused Decoding Enables 3D Anatomical Detection by Transformers
In this work, the Menze group proposes a novel Detection Transformer for 3D anatomical structure detection, dubbed Focused Decoder. Focused Decoder precisely focuses on relevant anatomical structures, using anatomical region atlas information to deploy query anchors, while at the same time restricting the cross-attention’s field of view to regions of interest. Evaluated on two publicly available CT datasets, Focused Decoder is shown to provide both strong detection results, as well as highly intuitive explainability via attention weights.
See Wittmann et al.,Journal of Machine Learning for Biomedical Imaging
Code is available at https://github.com/bwittmann/transoar -

Michael Krauthammer co-manages the establishment of the LOOP Zurich's new BioMedical Informatics Platform
The LOOP Zurich is establishing a central health data platform to enable FAIR data exchange between UZH, ETHZ and the four university hospitals in Zurich. Michael Krauthammer is the co-project manager of this new BioMedical Informatics Platform (BMIP), together with Prof. Gunnar Rätsch from ETHZ.
Read more about the new BioMedical Informatics Platform in UZH News: "Unlocking the Data Treasure Chest"
-

The Krauthammer lab and collaborators publish PRIDICT: An attention-based bidirectional recurrent neural network to predict prime editing efficiency and product purity
Predicting prime editing efficiency and product purity by deep learning
In this work, the Krauthammer lab and collaborators conducted a high-throughput screen to analyze prime editing outcomes of >90K pegRNAs on a set of >13K human pathogenic mutations. This dataset yielded sequence context features that influence prime editing and was subsequently used to train PRIDICT; an attention-based bidirectional recurrent neural network. PRIDICT reliably predicts editing rates for all small-sized genetic changes. Validation of PRIDICT, both on endogenous editing sites as well as on an external dataset, showed that pegRNAs with high versus low PRIDICT scores showed increased prime editing efficiencies in different cell types in vitro (12-fold) and in hepatocytes in vivo (tenfold), highlighting the value of PRIDICT for basic and translational research applications. PRIDICT is freely accessible at www.pridict.it.
See Mathis, Allam et al., Nature Biotechnology
This work is featured on UZH News: Artificial Intelligence Improves Efficiency of Genome Editing
-

The Bodenmiller group publishes a single-cell map of T cell exhaustion-associated immune environments in breast cancer
A comprehensive single-cell map of T cell exhaustion-associated immune environments in human breast cancer
Immune checkpoint therapy aims at preventing or reversing exhausted T cell states, but in breast cancer, T cell exhaustion is poorly understood. In this work, the Bodenmiller group used single-cell transcriptomics combined with imaging mass cytometry to systematically study immune environments of human luminal breast tumors with and without exhausted T cells. The data show that expression of PD-1 and CXCL13 on T cells, and MHC-I – but not PD-L1 – on tumor cells on tumor cells are powerful differentiators between these environments.
See Tietscher et al., Nat Communications -

The Polymenidou group and collaborators systematically evaluate human-derived anti-poly-GA antibodies in C9orf72 disease models
Comprehensive evaluation of human-derived anti-poly-GA antibodies in cellular and animal models of C9orf72 disease
Hexanucleotide G4C2 repeat expansions in the C9orf72 gene are the most common genetic cause of amyotrophic lateral sclerosis (ALS) and frontotemporal dementia (FTD). Dipeptide repeat proteins (DPRs) generated by translation of repeat-containing RNAs are key targets for therapeutic intervention. In this work, the Polymenidou group and collaborators generated human antibodies that bind DPRs with high affinity and specificity, systematically characterized these against multiple DPR species and tested the biological effects of antibodies targeting poly-GA in different cellular and mouse models.
See Jambeau et al, PNAS
-

The Menze group and collaborators publish the liver tumor segmentation benchmark
The Liver Tumor Segmentation Benchmark (LiTS)
In this work, the Menze group and collaborators report the set-up and results of the Liver Tumor Segmentation Benchmark (LiTS), which was organized in conjunction with the IEEE International Symposium on Biomedical Imaging (ISBI) 2017 and the International Conferences on Medical Image Computing and Computer-Assisted Intervention (MICCAI) 2017 and 2018. There was not a single algorithm that performed best for both liver and liver tumors in these three events. The best liver segmentation algorithm achieved a Dice score of 0.963, whereas the best algorithms for tumor segmentation achieved Dices scores of 0.674 (ISBI 2017), 0.702 (MICCAI 2017), and 0.739 (MICCAI 2018). Retrospectively, additional analysis on liver tumor detection revealed that not all top-performing segmentation algorithms worked well for tumor detection: The best liver tumor detection method achieved a lesion-wise recall of 0.458 (ISBI 2017), 0.515 (MICCAI 2017), and 0.554 (MICCAI 2018), indicating the need for further research. Nonetheless, LiTS remains an active benchmark and resource for research. See Bilic et al., Med Image Anal.
Both data and online evaluation are accessible via https://competitions.codalab.org/competitions/17094. -

The Bodenmiller group develops a strategy that estimates tissue spatial segregation to optimise multiplexed imaging experimental design
Optimizing multiplexed imaging experimental design through tissue spatial segregation estimation
Multiplexed imaging methods allow simultaneous detection of dozens of proteins and hundreds of RNAs, but parameters for the design of optimal multiplex imaging studies are lacking. In this work, the Bodenmiller group developed a statistical framework that determines the number and area of fields of view necessary to accurately identify all cell phenotypes that are part of a tissue. This strategy was then used on imaging mass cytometry data to identify a measurement of tissue spatial segregation that enables optimal experimental design. See Bost et al., Nat Methods
-

DQBM's roundup of 2022
Founded in 2019, the DQBM's mission is to foster research and education at the interface of biomedical research, biotechnology, and computational biomedicine, to develop the foundations of next-generation precision medicine. Ultimately, our goal is to advance precision medicine for the benefit of patients. To fulfil our mission, we rely on excellent science done by outstanding scientists. Here, we highlight the DQBM events and academic achievements in 2022.
-

The Polymenidou group's research is featured in EU GrantsAccess' Science Stories
A new perspective on neurodegenerative diseases
Read EU GrantsAccess' Science Stories feature on the Polymenidou group's research.
-

The Kümmerli group shows that collective decision-making in P. aeruginosa involves transient segregation of quorum-sensing activities across cells
Collective decision-making in Pseudomonas aeruginosa involves transient segregation of quorum-sensing activities across cells
Bacterial groups can coordinate cooperative actions through a communication process called quorum sensing (QS). However, whether individual bacteria coordinate their QS actions at the single-cell level, or whether group QS phenotypes are the sum of their noisy members, is unknown. Here, the Kümmerli group tracked the temporal commitments of individual Pseudomonas aeruginosa bacteria to the intertwined Las and Rhl-QS systems, from low to high population density. QS gene expression was shown to be noisy, with heterogeneity peaking during the build-up phase of QS. Discrete subgroups of cells formed that transiently segregated into two gene expression states: low Las-receptor expressers and high Las-receptor expressers. Over time, gene expression activities converged with all cells fully committing to QS. Using general mathematical models, the Kümmerli group was able to show that molecular resource limitations during the initiation phase of regulatory cascades can mechanistically trigger gene expression segregation across cells. This mechanism can operate as a built-in brake enabling a temporary bet-hedging strategy in unpredictable environments. Together, this work reveals that studying the behavior of bacterial individuals is key to understanding emergent collective actions at the group level.
See Jayakumar et al., Current Biology
-

Michael Krauthammer and collaborators publish a perspective paper on the use and ethics of Digital Twins in medicine
The Use and Ethics of Digital Twins in Medicine
This work introduces both the concept and use cases of digital twins in medicine, frames the debate on their ethical, legal and societal implications through the lens of related health digital technologies, machine learning and personalized medicine, and maps ethical challenges stemming from those. Finally, the authors lay out how digital twins may change and challenge the future practice of medicine.
See Iqbal, Krauthammer and Biller-Andorno, The Journal of Law, Medicine & Ethics
-

The Menze group publishes a new way of solving the inverse problem for brain tumor modeling
Learn-Morph-Infer: A new way of solving the inverse problem for brain tumor modeling
Current treatment planning of patients diagnosed with a brain tumor could benefit from assessment of the spatial distribution of tumor cell concentration. Magnetic resonance imaging (MRI) can reveal areas of high cell density in gliomas, but areas of low cell concentration, which can serve as a source for the secondary appearance of the tumor after treatment, are not detected. To estimate tumor cell densities beyond the visible boundaries of the lesion, numerical simulations of tumor growth could complement imaging information by providing estimates of full spatial distributions of tumor cells. In this work, the Menze group introduces Learn-Morph-Infer: a deep learning based methodology for inferring the patient-specific spatial distribution of brain tumors from T1Gd and FLAIR MRI medical scans. The method achieves real-time performance in the order of minutes on widely available hardware and the compute time is stable across tumor models of different complexity. As such, the proposed inverse solution approach allows for clinical translation of brain tumor personalization and could also be adopted to other scientific and engineering domains.
See Ezhov et al., Med Image Anal.
-

The Kümmerli group shows that loss-of-function and regulon modulation drives diversification in quorum sensing activity patterns in P. aeruginosa
Evolution of quorum sensing in Pseudomonas aeruginosa can occur via loss of function and regulon modulation
Pseudomonas aeruginosa uses quorum sensing (QS) to coordinate expression of traits required for growth and virulence in the context of infections. Despite its importance for bacterial fitness, the QS regulon appears to be a common mutational target during long-term adaptation of P. aeruginosa in the host, natural environments, and experimental evolutions. By examining mutation types in the three QS regulons of 61 experimentally evolved QS mutants, the Kümmerli group found that mutations involving the master regulator, LasR, resulted in an almost complete breakdown of QS, whereas mutations in RhlR and PqsR resulted in changes in regulon structure and the QS-regulated trait profile. Beyond affecting the plasticity and diversity of evolved populations, these mutations might also impact bacterial fitness and virulence during infections.
See Jayakumar, Figueiredo & Kümmerli, mSystems
-

The Menze group improves deep learning based super-resolution of 4D-flow MRI data
SRflow: Deep learning based super-resolution of 4D-flow MRI data
The quantification of hemodynamics using 4D-flow magnetic resonance imaging (MRI) data requires an adequate spatio-temporal vector field resolution at a low noise level. Here, the Menze group provides a deep convolutional neural network (CNN) that learns the inter-scale relationship of the velocity vector map and leverages an efficient residual learning scheme to make it computationally feasible. A detailed comparative study between the proposed super-resolution and the conventional cubic B-spline based vector-field super-resolution shows that the new method not only improves the peak-velocity to noise ratio of the flow field by 10% and 30% for in vivo cardiovascular and cerebrovascular data, respectively, for 4 × super-resolution, but also offers 10x faster inference over the state-of-the-art.
See Shit et al., Front. Artif. Intell
-

Magdalini Polymenidou granted a Target ALS and Alzheimer’s Drug Discovery Foundation Award for Biomarker Research
We congratulate Prof. Dr. Magdalini Polymenidou and her lab for receiving the Target ALS and Alzheimer’s Drug Discovery Foundation Award for Biomarker Research, for their collaborative project with the University of Verona and Trieste International School for Advanced Studies. The consortium will work towards the development of a diagnostic test for TDP-43 proteinopathies using nasal swabs.
-

The Krauthammer lab is part of the newly funded DIZH innovation structure "Zurich Applied Digital Health Center"
Michael Krauthammer is core team member of the newly funded DIZH innovation structure Zurich Applied Digital Health Center: a practice lab for patient-centered clinical innovation.
-

The Kümmerli group shows coordination of siderophore gene expression in clonal P. aeruginosa cells
Coordination of siderophore gene expression among clonal cells of the bacterium Pseudomonas aeruginosa
An open question in microbiology is whether seemingly coordinated group-level responses, such as biofilm formation, actually mirror what individual cells do. To tackle this question, the Kümmerli group used single-cell microscopy to simultaneously quantify the investment of individual P. aeruginosa cells into two public goods. Using gene expression as a proxy for investment, these bacterial cells initially show no coordination in siderophore investment, but rather high heterogeneity and bi-modality. However, with increasing cell density, gene expression becomes more homogenised across cells, with positive associations in siderophore gene expression across cells and with cell-to-cell variation correlating with cellular metabolic states. This suggests that siderophore-mediated signalling aligns behavior of individual bacteria over time and spurs a coordinated three-phase siderophore investment cycle, covering the time spans from low to high population density, steered by the various interconnected regulatory mechanisms governing siderophore synthesis.
See Mridha & Kümmerli, Commun. Biol.
-

The Bodenmiller group's research is featured in EU GrantsAccess' Science Stories
With the Three-Dimensional Cell Atlas to Precision Medicine
Read EU GrantsAccess' Science Stories feature on the Bodenmiller group's research.
-

The Joller group publishes a review on the interplay between regulatory T cells and peripheral tissues
Moving to the Outskirts: Interplay Between Regulatory T Cells and Peripheral Tissues
Regulatory T cells (Tregs) curb excessive immune responses and dampen inflammation. Furthermore, in non-lymphoid tissues, Tregs promote tissue homeostasis, regeneration and repair. Profound understanding of the tissue-specific adaptations and functions of these Tregs might pave the way for therapeutic approaches targeting their regenerative role. In this review, the Joller group outlines their current understanding of how Tregs migrate into peripheral tissues and the factors required for their maintenance at these sites. Furthermore, tissue-specific adaptations of Tregs at barrier and immuno-privileged sites are discussed, as well as the mechanisms that regulate Tregs' function within these organs. Finally, this work outlines what is known about the interactions of Tregs with non-immune cells in the different peripheral tissues at steady state and upon challenge or tissue damage.
See Estrada Brull, Panetti & Joller, Frontiers in Immunology
-

The Menze group develops a convolutional neural network for residual motion correction in fast whole-brain MRI
Learning residual motion correction for fast and robust 3D multiparametric MRI
In routine clinical quantitative magnetic resonance imaging (MRI), motion artifacts affect parameter estimation and thus data quality. In this work, the Menze group presents a multiscale 3D convolutional neural network (CNN) that learns the nonlinear relationship between motion-influenced quantitative parameter maps and the residual error to their motion-free reference. A physically informed simulation is proposed for supervised model training, which generates independent paired data sets from a priori motion-free data. The proposed motion correction CNN outperforms the current state-of-the-art and reliably provides high, clinically relevant image quality for mild to pronounced patient motion.
See Pirkl et al., Med. Image Anal.
-

The Kümmerli group shows that enforced specialization fosters mutual cheating, but not division of labour, in P. aeruginosa
Enforced specialization fosters mutual cheating and not division of labour in the bacterium Pseudomonas aeruginosa
An open question in microbiology is whether natural selection can favour division of labour where subpopulations or species specialise in the production of a single public good, whilst sharing the complementary goods at the group level. In this work, the Kümmerli group explored the conditions under which specialisation can lead to division of labour. By growing engineered specialists of the bacterium P. aeruginosa, each of which which only produce one of two siderophores, at different mixing ratios under varying levels of iron limitation, they could show that enforcing specialisation with regard to siderophore production does not lead to beneficial division of labour in P. aeruginosa. Rather, this enforced specialisation leads to the stable co-existence of the two specialists through mutual cheating. Mridha and Kümmerli propose that in generalists, natural selection might favour fine-tuned regulatory mechanisms over division of labour, because in fluctuating environments, this fine-tuning allows generalists to maintain the flexibility to adequately adjust public good investments.
See Mridha & Kümmerli, J Evol Biol.
-

The Bodenmiller group characterises chemokine expression and function in melanoma
Multiplexed imaging mass cytometry of the chemokine milieus in melanoma characterizes features of the response to immunotherapy
To predict the effects of immunotherapy in cancer, better spatial understanding of the tumor microenvironment (TME) is needed. In this work, the Bodenmiller group characterised chemokine expression and function in samples from 69 patients with metastatic melanoma. Using multiplexed mass cytometry-based imaging of protein markers and RNA transcripts, they found that CXCL9 and CXCL10 were present in patches with CXCL13+ exhausted T cells, suggesting that they recruited B cells and aided in the formation of tertiary lymphoid structures (TLS) in melanoma. TLS had a spatial enrichment of naïve and naïve-like T cells, which are involved in anti-tumor responses. Together, this study highlights the strength of targeted RNA and protein co-detection to analyse TME based on chemokine expression and suggests that the formation of tertiary lymphoid structures may be accompanied by naïve and naïve-like T cell recruitment, which may contribute to anti-tumor activity.See Hoch, Schulz et al., Sci Immunol
-

Magdalini Polymenidou receives an ERC Consolidator Grant
We congratulate Prof. Dr. Magdalini Polymenidou, who has been awarded the prestigious ERC Consolidator Grant for the project “TDP-43 transitions”.
Please have a look at the UZH News and ERC Announcement
-

The Menze group develops a deep neural network to reconstruct the 3D standing spine posture from 2D Radiographs
Anatomy-Aware Inference of the 3D Standing Spine Posture from 2D Radiographs
In various spinal disorders, a biomechanical load analysis of the spine in the upright position is useful to understand the underlying causes of the disorder and to help guide therapy. Despite the complex 3D shape of the human spine, this analysis is typically performed using 2D radiographs. Here, the Menze group proposes a novel deep neural network architecture, which takes orthogonal 2D radiographs and infers the spine’s 3D posture using vertebral shape priors to reconstruct the 3D spinal pose in an upright standing position.
See Bayat et al., Tomography
-

The Polymenidou group publishes a protocol for the identification of RNA–RBP interactions in subcellular compartments by CLIP-Seq
Cross-linking immunoprecipitation and high-throughput sequencing (CLIP-seq) allows the identification of RNA bound by RNA-binding proteins (RBPs) in vivo and ex vivo with high specificity. In this protocol paper, the Polymenidou lab describe the adaptation of CLIP-seq to identify the specific RNA targets of an RBP (FUS) at neuronal synapses, including subcompartment isolation, RBP–RNA complex enrichment, and upscaling steps.
See Sahadevan, Pérez-Berlanga & Polymenidou,Methods Mol. Biol.
-

Zsolt Balázs (Krauthammer group) wins the Pfizer Research Prize 2022
We congratulate Dr. Zsolt Balázs (Krauthammer group) for winning the Pfizer Research Prize 2022 together with Dr. Egle Ramelyte and Dr. Aizhan Tastanova (University Hospital Zurich) for their single-cell RNA studies on the effects of oncolytic viral therapy in cutaneous lymphoma. (Ramelyte et al., Cancer Cell).
Please find the full announcement here: Stiftung Pfizer Forschungspreis
and the UZH News article here.
-

Watch Bernd Bodenmiller's video on innovative research at the Comprehensive Cancer Center Zurich (german only)
In diesem Video erklärt Bernd Bodenmiller, wie die innovative Krebsforschung am Comprehensive Cancer Center Zurich (CCCZ) Krebspatientinnen und -patienten Zugang zu personalisierten Behandlungskonzepten ermöglicht. -

The Kümmerli group and collaborators publish a high-throughput method to elucidate siderophores from natural Pseudomonas
A comprehensive method to elucidate pyoverdines produced by fluorescent Pseudomonas spp. by UHPLC-HR-MS/MS
This work resulted from a collaboration with the research group of Prof. Laurent Bigler at the University of Zurich's Institute for Chemistry.
Microorganisms produce secondary metabolites, which are low molecular weight compounds with various bioactive properties that play key roles in microbial physiology, growth and survival. In recent years, interest in microbial secondary metabolites has surged, owing to their potential as a source for drug discovery, as well as their application in agriculture and as biosurfactants. This work presents a novel UHPLC-MS/MS-based method for the high-throughput characterisation of the secondary metabolites pyoverdines from crude bacterial extracts.See Rehm et al, Analytical and Bioanalytical Chemistry
-

The Bodenmiller group develops a 3D IMC approach for multiplexed 3D tissue analysis at single-cell resolution
Three-dimensional imaging mass cytometry for highly multiplexed molecular and cellular mapping of tissues and the tumor microenvironment
Understanding tissue function and pathology requires knowledge of the molecular components in their original three-dimensional (3D) context. In this work, the Bodenmiller group reports on the development of 3D Imaging Mass Cytometry (IMC) for multiplexed 3D tissue analysis and demonstrates, using the analysis of human breast cancer samples, that the detailed models generated by 3D IMC enable comprehensive analysis of the cellular microenvironment and tissue architecture at single-cell resolution. Thus, 3D-IMC advances the study of the spatial distribution of proteins in the context of 3D tissue architecture and will prove powerful in the study of phenomena such as tumor cell invasion.
See Kuett et al., Nature Cancer
-

The Menze group publishes a geometry-aware neural solver for fast Bayesian calibration of brain tumor models
Geometry-aware neural solver for fast Bayesian calibration of brain tumor models
Current modeling approaches for brain tumor dynamics, based on numerical solvers that simulate tumor growth using a given differential equation, are too time-consuming for clinical implementation. Recent data-driven approaches are able to emulate physical simulations, but typically fail to generalise across the variability of boundary conditions imposed by patient-specific anatomy. Here, the Menze group proposes a learnable surrogate for simulating tumor growth that maps biophysical model parameters directly to simulation results, while taking patient geometry into account. This neural solver is tested in a Bayesian model personalisation task for a cohort of glioma patients. Bayesian inference using the proposed surrogate leads to estimates analogous to those obtained by solving the forward model using a regular numerical solver. Owing to the near real-time computational cost, however, the method proposed here is suitable for clinical settings.See Ezhov et al., IEEE Trans Med Imaging
Access the code via: https://github.com/IvanEz/tumor-surrogate
-

DQBM's roundup of 2021
Founded in 2019, the DQBM's mission is to foster research and education at the interface of biomedical research, biotechnology, and computational biomedicine, to develop the foundations of next-generation precision medicine. Ultimately, our goal is to advance precision medicine for the benefit of patients.
-

Michael Krauthammer's team is part of the new Innosuisse Flagship Project 'Smart Hospital'
To create solutions for the hospital of the future, Michael Krauthammer and his team will contribute to the new Innosuisse Flagship Project 'Smart Hospital - Integrated Framework, Tools & Solutions'. This collaborative project focuses on new, holistic organisational approaches and digital technologies.
-

Prof. Dr. Nicole Joller and her team join the DQBM
The DQBM is delighted to announce that Prof. Dr. Nicole Joller will join the DQBM Faculty as Associate Professor for Immunology in January 2022.
Prof. Joller studied Biochemistry at ETH Zurich, where she obtained her PhD in 2008. Subsequently, she was a Postdoctoral Research Fellow at Brigham and Women's Hospital, Harvard Medical School and the Broad Institute at MIT and Harvard, US. In 2014, Prof. Joller joined UZH as Assistant Professor at the Institute of Experimental Immunology.
Please find the UZH's official announcement of Prof. Joller's appointment as Associate Professor for Immunology here (German only):
https://www.media.uzh.ch/de/universitaetsrat/2021/Dezember.htmlPlease also visit Nicole Joller's group website
We look forward to welcoming the Joller Group to our Department next month!
-

Rolf Kümmerli appointed Associate Professor for Evolution of Human Microbiomes and Pathogens
We congratulate Prof. Dr. Rolf Kümmerli on his appointment as Associate Professor for Evolution of Human Microbiomes and Pathogens, effective January 1, 2022.
Please see the official announcement here (in German only):
https://www.media.uzh.ch/de/universitaetsrat/2021/Dezember.html -

The Krauthammer group and collaborators publish a perspective paper on analyzing patient trajectories with AI
Analyzing Patient Trajectories With Artificial Intelligence
Patient data recorded over time form unique patient histories that can predict future disease progression and thereby facilitate effective care. However, digital medicine often uses limited health events data from a single or small number of time points, ignoring additional information encoded in patient trajectories. This paper provides an overview of recent efforts to develop AI solutions that incorporate trajectories, with a particular focus on the implications for developing disease models from patient trajectories in the context of the typical AI workflow, and concludes with a discussion of how such AI solutions will enable the development of robust models for personalized risk assessment, subtyping, and disease pathway discovery.
See Allam et al., JMIR
-

The Polymenidou group is awarded an SNSF Sinergia Grant together with collaborators at UZH and ETHZ
Congratulations to Magdalini Polymenidou and her research group for being awarded an SNSF Sinergia grant in collaboration with Prof. Benjamin Schuler (UZH, Biochemistry), Prof. Frederic Allain (ETH, Biochemistry) and Prof. Gunnar Jenschke (ETH, Chemistry and Applied Biosciences) for the project: "Protein disorder in RNA-protein interactions: from dynamic structures to pathology".
-

"Bridging the Gap to Patients"
On the occasion of the DQBM Founding Symposium: Frontiers in Biomedical Research, UZH News interviewed DQBM Founding Director Bernd Bodenmiller. Please find the article here
-

The Polymenidou group shows that patient-derived pathological TDP-43 triggers de novo aggregation of physiological TDP-43 in host cells
FTLD-TDP assemblies seed neoaggregates with subtype-specific features via a prion-like cascade
Frontotemporal dementia (FTD) is characterised by frontotemporal lobar degeneration (FTLD) and accumulation of pathological protein aggregates. FTLD-TDP subtypes are characterised by the accumulation and aggregation of the RNA-binding protein TDP-43. Notably, clinically different FTLD-TDP subtypes show morphologically distinct TDP-43 aggregates. However, the mechanism of the emergence of these aggregates and their contribution to clinical heterogeneity are poorly understood. One crucial question is whether pathological TDP-43 follows a prion-like cascade of amplification and template-directed conversion during disease propagation, and if so, which are the molecular determinants of this process. This study used advanced microscopy techniques to compare the seeding properties of pathological aggregates in two FTLD-TDP subtypes to show that pathological TDP-43 triggers template-dependent aggregation and amplification of neoaggregates. These pathological aggregates have different, subtype-specific seeding potency and follow distinct phosphorylation timelines. Moreover, neoaggregates triggered by different subtypes display different aggregation profiles, resembling the original aggregates in patient brains. Thus these cellular seeding models replicate aspects of the patient pathological diversity and will be a useful tool in the quest for subtype-specific therapeutics.
See De Rossi et al., EMBO Rep
-

Johanna Wagner (Alumnus Bodenmiller group) wins the 2021 Ida de Pottère-Leupold and Dr. iur. Erik de Pottère Cancer Research Award
We congratulate Dr. Johanna Wagner for winning the 2021 Ida de Pottère-Leupold and Dr. iur. Erik de Pottère Cancer Research Award. This award recognises her PhD work performed in the Bodenmiller group: A single-cell atlas of the tumor and immune ecosystem of human breast cancer (Wagner et al., Cell 2019).
Please find the full announcement here: CCCZ
-

The Kümmerli group identifies a dual role for siderophores in driving invasion dynamics in bacterial communities
Siderophores drive invasion dynamics in bacterial communities through their dual role as public good versus public bad
While microbial invasions are known to disturb ecosystems and incite dysbiosis and disease in hosts, the mechanisms determining invasion outcomes are often unclear. This study examined the role of iron-scavenging siderophores in driving invasions of Pseudomonas aeruginosa into resident communities of environmental pseudomonads. Through supernatant feeding and invasion assays, it was shown that invasion success increased when invaders could use their siderophores to inhibit resident bacterial growth, while invasion success decreased when invaders were inhibited by the residents’ siderophores. Thus, this work identifies iron-scavenging siderophores as a major driver of invasion dynamics in bacterial communities under iron-limited conditions.See Figueiredo et al, Ecol Lett
-

Johanna Furrer (Polymenidou group) wins the Faculty of Science's Semester Prize
We congratulate Johanna Furrer, MSc from the Polymenidou group for winning the Faculty of Science's Semester Prize for her MSc thesis: 'Developing new cellular models to explore TDP-43-associated pathologies'.
-

The Polymenidou group publishes a 'Spotlight' on benefits of distributing protein aggregates among microglial networks
Sharing is caring: The benefits of distributing protein aggregates among microglial networks
Trafficking of protein aggregates through neural networks is known to cause propagation of pathology and toxicity in neurodegenerative diseases. Scheiblich et al. (2021) describe an advantageous aggregate-sharing strategy, challenging the view that pathological protein propagation only has a detrimental outcome on network longevity.
Spotlight article: Wiersma and Polymenidou, Neuron
Original publication: Scheiblling et al, Cell
-

The Menze group publishes a deep learning-based approach to segment the claustrum in human brain MRI scans
Automated claustrum segmentation in human brain MRI using deep learning
Despite the central role of the claustrum in the structure and function of the mammalian forebrain, few in vivo human studies of this delicate, sheet-like structure exist. Here, the Menze group and their collaborators present a Deep Learning-based multi-view approach for segmenting the claustrum in T1-weighted MRI scans. The algorithm enables robust automatic claustrum segmentation and therefore holds potential for facilitating MRI-based human claustrum studies.
See Li et al., Hum Brain Mapp.
Access the software and models via: https://github.com/hongweilibran/claustrum_multi_view -

DQBM's wet lab groups are moving into UZI5
From 8-13 September, the DQBM wet lab groups (Bodenmiller, Kümmerli and Polymenidou) are moving into the new UZH lab building UZI5 on Campus Irchel.
-

The Kümmerli group uses single-cell imaging to show that S. aureus is highly competitive against P. aeruginosa on surfaces
Single-Cell Imaging Reveals That Staphylococcus aureus Is Highly Competitive Against Pseudomonas aeruginosa on Surfaces
In polymicrobial infections, interactions between bacterial species can complicate both disease progression and treatment options. Pseudomonas aeruginosa and Staphylococcus aureus often occur together in such infections and tend to colonize surfaces to form aggregates and biofilms. This study addressed whether and how individual cells interact with each other on solid substrates, rather than in batch cultures. Single-cell time-lapse fluorescence microscopy, combined with automated image analysis, showed that, although P. aeruginosa is generally considered dominant over S. aureus based on batch culture studies, on surfaces the competitive balance tips in favor of S. aureus. Thus, quantitative single-cell live imaging offers the potential to reveal microbial behaviors that cannot be predicted from batch culture studies, advancing our understanding of pathogen interactions on host-associated surfaces during polymicrobial infections.See Niggli, Wechsler & Kümmerli, Front. Cell. Infect. Microbiol
-

The Krauthammer group publishes BE-DICT: An attention-based deep learning algorithm to predict base editing outcomes
Predicting base editing outcomes with an attention-based deep learning algorithm trained on high-throughput target library screens
This work results from a collaboration with the research group of Prof. Dr. Gerald Schwank at the University of Zurich.
Base editors consist of a DNA-targeting CRISPR-Cas module and a single-stranded DNA deaminase. These chimeric ribonucleoprotein complexes enable transition of C•G into T•A base pairs and vice versa on genomic DNA. So far, base editors have not been widely used in research or therapy, due to broad variation in their editing efficiencies on different genomic loci. To overcome this hurdle, this study performed an extensive analysis of A- and C base editors on a library of 28,294 lentivirally integrated genetic sequences to establish BE-DICT, an attention-based deep learning algorithm capable of predicting base editing outcomes with high accuracy. The BE-DICT tool can in principle be trained on any novel base editor variant, facilitating the application of base editing for research and therapy.See Marquart, Allam, Janjuha et al., Nature Communications
The BE-DICT web app can be found here
-

The Krauthammer group publishes AttentionDDI: Siamese attention-based deep learning method for drug-drug interaction predictions
AttentionDDI: Siamese attention-based deep learning method for drug-drug interaction predictions
Drug-drug interactions (DDIs) can occur when two or more drugs are administered, leading to side effects beyond those observed when drugs are taken by themselves. As the number of possible drug pairs is enormous, it is nearly impossible to experimentally test all combinations and discover previously unobserved side effects. This is where machine learning methods can help.
This study proposes a Siamese self-attention multi-modal neural network for DDI prediction. AttentionDDI integrates multiple drug similarity measures derived from a comparison of drug characteristics, including drug targets, pathways and gene expression profiles. The proposed model is able to accurately predict DDIs and beneficially applies an Attention mechanism, typically used in the Natural Language Processing domain, to aid in DDI model explainability.
See Schwarz et al., BMC Bioinformatics -

The Kümmerli group shows that ecology drives evolution of social strategies in P. aeruginosa
Ecology drives the evolution of diverse social strategies in Pseudomonas aeruginosa
While much is known about the evolution of bacterial cheating resistance mechanisms and environmental factors that maintain cooperation when cheaters are present, the evolution of cooperative traits in environments where cheating is insignificant is less understood. In this study, an experimental evolution setup was used to follow changes in the production of the public good pyoverdine, an iron-scavenging siderophore of P. aeruginosa. 1200 generations of bacterial evolution in nine different environments revealed that social traits can evolve rapidly in different directions, with particularly strong selection for higher levels of cooperation in environments where individual dispersal is lower, as predicted by social evolution theory. Moreover, a regulatory link is established between pyoverdine production and quorum-sensing, showing that increased cooperation with respect to one social trait can be associated with the loss of another trait.
See Figueiredo, Wagner & Kümmerli, Molecular Ecology
N.B. This article has been accepted for publication and undergone full peer review but has not been through the copyediting, typesetting, pagination and proofreading process, which may lead to differences between this version and the Version of Record. -

The Polymenidou group and collaborators show that LAG3 does not modulate α-synucleinopathies
LAG3 is not expressed in human and murine neurons and does not modulate α-synucleinopathies
Recently, it was proposed that neuronal lymphocyte-activation gene 3 (LAG3) may function in the central nervous system as a receptor of pathogenic α-synuclein assemblies, which are causally involved in Parkinson’s disease. This study investigated this putative role of the LAG3 immune checkpoint molecule in the spread of α-synucleinopathies by analysing the expression pattern of LAG3 in human and mouse brains. Despite using various methods and model systems, no evidence was found for LAG3 expression in neurons. While the interaction of LAG3 with α-synuclein fibrils was confirmed, the specificity of this interaction appeared limited. Moreover, overexpression of LAG3 in cultured human neural cells did not cause increased deposition of α-synuclein aggregates ex vivo and overall survival of human A53T α-synuclein transgenic mice was unaffected by LAG3 depletion. Finally, the seeded induction of α-synuclein lesions in hippocampal slice cultures was unaffected by LAG3 knockout. Overall, this study indicates that the proposed role of LAG3 in the propagation of α-synucleinopathies is not universally valid and that the search for relevant targets to slow or completely abolish the pathogenesis of neurodegenerative diseases must continue.See Emmenegger et al,. EMBO Mol Med.
-

The Menze group and collaborators publish VerSe: A vertebrae labelling and segmentation benchmark for multi-detector CT images
VerSe: A vertebrae labelling and segmentation benchmark for multi-detector CT images
Reliable and accurate automated processing of spine images is expected to benefit clinical decisions on diagnosis and surgery planning, as well as population-based analysis of spine and bone health. The 2019 and 2020 Large Scale Vertebrae Segmentation Challenge (VerSe), organised in conjunction with the International Conference on Medical Image Computing and Computer Assisted Intervention (MICCAI), therefore called for algorithms tackling the labeling and segmentation of vertebrae. This work presents the benchmarking of 25 algorithms on two datasets containing 374 multi-detector CT scans from 355 patients, from which 4505 vertebrae were individually annotated at voxel level by a human-machine hybrid algorithm. Performance variation was investigated at the vertebra- and scan level, as well as for different fields of view. Furthermore, the generalisability of approaches was assessed by evaluating the top-performing algorithms of one challenge iteration on data from the other iteration.
See Sekuboyina et al.,Medical Image Analysis
Access the VerSe content and code via: https://github.com/anjany/verse. -

Bjoern Menze and Ender Konukoglu (ETHZ) organise 'MIDL 2022 - Medical Imaging with Deep Learning' in Zurich
MIDL 2022 will take place from 6-8 July 2022 and is hosted by UZH and ETHZ
MIDL 2022 is organised by Bjoern Menze (DQBM) and Ender Konukoglu (ETHZ). We are looking forward to welcoming everyone to the charming city as well as stimulating a fruitful scientific exchange.
-

Watch the #NCCRWomen feature on Magdalini Polymenidou
Please find the #NCCRWomen feature on Magdalini Polymenidou, DQBM Professor and part of the NCCR RNA & Disease here.
-

The Kümmerli group publishes their work on drivers of siderophore-mediated social interactions in natural bacterial communities
Local adaptation, geographical distance and phylogenetic relatedness: assessing the drivers of siderophore-mediated social interactions in natural bacterial communities
This study tested whether social microbe-microbe interactions can spur patterns of local adaptation in bacterial populations. To this end, the production of siderophores was assessed in soil and pond communities of Pseudomonas bacteria, collected across different geographical scales. While supernatant-feeding and competition assays revealed large variation in the extent to which 'unsocial' bacteria (cheaters) can benefit from 'social' ones (cooperators), this variation depended primarily on phylogenetic relatedness, and not on geographical distance. Thus, common ancestry - rather than geographical distance - is the main predictor of siderophore-mediated social interactions among Pseudomonas bacteria.
See Butaitė, Kramer & Kümmerli, J Evol Biol
N.B. This article has been accepted for publication and undergone full peer review but has not been through the copyediting, typesetting, pagination and proofreading process, which may lead to differences between this version and the Version of Record. -

The Polymenidou group shows that synaptic FUS accumulation triggers early misregulation of synaptic RNAs in an ALS mouse model
Synaptic FUS accumulation triggers early misregulation of synaptic RNAs in a mouse model of ALS
This study investigates a molecular mechanism triggering early dysfunction in neurons of patients suffering from a familial form of amyotrophic lateral sclerosis (ALS), linked to mutations in the RNA-binding protein FUS. In this work, the Polymenidou team uncovers that the protein FUS plays a novel role in regulating the levels of specific RNAs at the synapses, which are the small and critical points of communication between neurons. The team deciphered the direct molecular interactions between FUS protein and synaptic RNAs and showed that this FUS-mediated dysregulation occurs very early, prior to the development of motor symptoms in ALS.
See Sahadevan, Hembach et al, Nature Communications.
This paper was published back-to-back with Scekic-Zahirovic, Sanjuan-Ruiz et al, Nature Communications. -

Watch Michael Krauthammer's Inaugural Lecture 'Wie Daten und künstliche Intelligenz die Medizin von morgen prägen'
Please find the recording of Michael Krauthammer's Inaugural Lecture Wie Daten und künstliche Intelligenz die Medizin von morgen prägen here (start at 1:59:20).
-

The Bodenmiller group shows that mapping the breast cancer signaling network improves drug sensitivity prediction
Deciphering the signaling network of breast cancer improves drug sensitivity prediction
This work provides proof of principle that pesonalized single-cell measurements combined with mechanistic modeling could guide effective precision medicine strategies. Using mass cytometry, the Bodenmiller group characterised signaling landscapes in 62 breast cancer cell lines and five healthy tissue cell lines. Data from over 80 million single cells under 4,000 conditions were used to build cell line-specific signaling network models that both accurately predicted drug sensitivity and identified genomic features associated with drug sensitivity, as validated in patient-derived xenograft mouse models.
See Tognetti et al., Cell Systems
-

Watch Michael Krauthammer's 'Clinics meets Bioinformatics' Keynote on AI Support in the Clinic
Michael Krauthammer gave a Keynote on 'AI Support in the Clinic' at the Comprehensive Cancer Center Zurich (C3Z)'s 'Clinics Meets Bioinformatics' Symposium. Please find the recordinghere.
More about Comprehensive Cancer Center Zurich -

The Bodenmiller group publishes their collaborative work on dysregulation of T cell homeostasis and function in severe COVID-19
Profound dysregulation of T cell homeostasisand function in patients with severe COVID-19
This work results from a collaboration between the groups of Bernd Bodenmiller and Jakob Nilsson and Onur Boyman at the Department of Immunology, University Hospital Zurich (USZ).
See Adamo et al., Allergy
N.B. This article has been accepted for publication and undergone full peer review but has not been through the copyediting, typesetting, pagination and proofreading process, which may lead to differences between this version and the Version of Record. -

The Krauthammer group publishes their collaborative work on AI support for ethical decision-making around resuscitation
AI support for ethical decision-making around resuscitation: proceed with care
This article discusses the results of an interview study with healthcare professionals at a university hospital. The study aimed to understand the status quo of decision making processes around resuscitation and to explore a potential role for AI systems in decision-making around code status.
This work results from a collaboration between the Krauthammer lab (UZH & USZ) and colleagues from (i) the Institute of Biomedical Ethics and History of Medicine (UZH), (ii) the Department of Management, Technology, and Economics (ETHZ), (iii) Clinical Ethics (USZ) and (iv) the Computational Science and Engineering Laboratory (ETHZ).
See Biller-Andorno et al., Journal of Medical Ethics -

The Menze group publishes their review on deep learning for medical image analysis
Deep learning for medical image analysis: a brief introduction
Progress in the field of deep learning has led to the development of neural network algorithms that can compete with, or even surpass, human performance in vision tasks, such as image classification or segmentation. This review provides a brief introduction to the building blocks of neural networks and explains why convolutional neural networks (CNNs) in particular excel at identifying relevant image features. In addition, it discusses how these CNNs learn to associate relevant image features with clinical features of interest, and how CNNs automatically segment structures of interest in an image volume. Finally, obstacles for wider application of these algorithms in scientific and clinical practice are addressed.
See Wiestler & Menze, Neuro-Oncology Advances
-

The Krauthammer group publishes their collaborative work on oncolytic virotherapy-mediated anti-tumor response
Oncolytic virotherapy-mediated anti-tumor response: a single-cell perspective
This work results from a collaboration between the Krauthammer lab and the research groups of Mitchell P. Levesque and Reinhard Dummer at the Dermatology Department, University Hospital Zurich and UZH.
See Ramelyte, Tastanova, Balázs et al., Cancer Cell.
-

The Tumor Profiler Consortium publishes their study on integrated, multi-omic, functional tumor profiling for clinical decision support
The Tumor Profiler Study: integrated, multi-omic, functional tumor profiling for clinical decision support
By integrating data from innovative molecular profiling technologies, new opportunities for Precision Medicine arise. The Tumor Profiler Study is a multi-centre, multi-method observational trial. It combines a prospective diagnostic approach that probes the relevance of in-depth tumor profiling for clinical decision-making, with an exploratory approach to improve our biological understanding of the disease. In this highly collaborative project, the Bodenmiller lab contributes its expertise in single-cell CyTOF and imaging mass cytometry (IMC).
See Irmisch et al., Cancer Cell.
Read more about the Tumor Profiler Project in UZH news.
-

The Bodenmiller group publishes the cytomapper package, a tool to visualise highly multiplexed imaging data in R
Cytomapper: an R/bioconductor package for visualisation of highly multiplexed imaging data
In this work, the Bodenmiller group describes the R package "cytomapper", which enables visualisation of pixel- and cell-level information obtained by multiplexed imaging.
See Eling et al., Bioinformatics.
-

The Bodenmiller group publishes their collaborative work on innate immune responses in COVID-19
A distinct innate immune signature marks progression from mild to severe COVID-19
This work results from a collaboration between the groups of Bernd Bodenmiller and Jakob Nilsson and Onur Boyman at the Department of Immunology, University Hospital Zurich (USZ).
See Chevrier et al., Cell Rep. Medicine.
-

The Polymenidou group publishes their collaborative work on direct binding of importins to arginine-rich dipeptide repeat proteins
Nuclear Import Receptors Directly Bind to Arginine-Rich Dipeptide Repeat Proteins and Suppress Their Pathological Interactions
This work results from a collaboration between the groups of Magdalini Polymenidou and J. Shorter (Perelman School of Medicine, University of Pennsylvania, Philadelphia USA), D. Edbauer (German Center for Neurodegenerative Diseases (DZNE), Munich, Germany), T. Madl (Medical University of Graz, Austria) and D. Dormann (Ludwig-Maximilians-University Munich, Germany).
See Hutten et al, Cell Reports -

DQBM's roundup of 2020
Founded in 2019, the DQBM's mission is to foster research and education at the interface of biomedical research, biotechnology, and computational biology, to develop the foundations of next-generation precision medicine. Ultimately, our goal is to advance precision medicine for the benefit of patients.
-

The Bodenmiller group publishes their work on spatial phenotypic variability in 3D spheroids
A quantitative analysis of the interplay of environment, neighborhood, and cell state in 3D spheroids
In this work, the Bodenmiller group developed a novel barcoding approach that enables high throughput multiplexed imaging of 3D microtissues using Imaging Mass Cytometry.
See Zanotelli et al., Mol Syst Biol.
-

The Kümmerli group publishes their collaborative work on population invasion through exploitation of Pseudomonas aeruginosa public goods
Loss of a pyoverdine secondary receptor in Pseudomonas aeruginosa results in a fitter strain suitable for population invasion
This work results from a collaboration between the groups of Rolf Kümmerli and José I. Jiménez at the Department of Life Sciences, Imperial College London.
The emergence of antibiotic resistant bacterial pathogens in healthcare requires the development of novel treatment strategies. Promising avenues include the exploitation of microbial social interactions based on public goods, which are produced by cooperative microorganisms at a fitness cost and can be exploited by cheaters that do not produce these goods. In this work, González and colleagues took a novel approach by manipulating uptake (rather than production) of public goods, focusing on the siderophore pyoverdine produced by the human pathogen Pseudomonas aeruginosa. Deletion and/or overexpression of the pyoverdine primary and secondary receptors revealed that receptor synthesis feeds back on the rates of pyoverdine production and uptake, resulting in bacterial strains with altered pyoverdine-associated costs and benefits. These findings show that manipulation of public good uptake can result in fitter bacterial strains suitable for population invasion, which could potentially be harnessed for medical interventions.See González et al, ISME Journal
-

The Menze group publishes DeepVesselNet, a deep learning approach to perform vessel segmentation, centerline prediction, and bifurcation detection tasks
DeepVesselNet: Vessel Segmentation, Centerline Prediction, and Bifurcation Detection in 3-D Angiographic Volumes
See Tetteh et al, Front. Neurosci.
-

The Menze group publishes a Deep Learning-enabled tool for segmentation of organs in whole-body mouse scans
Deep learning-enabled multi-organ segmentation in whole-body mouse scans
The Menze lab presents a deep learning solution called AIMOS that automatically segments major organs and the skeleton in whole-body images of mice. AIMOS is a powerful open-source solution that increases scalability, reduces bias, and fosters reproducibility in biomedical research, as illustrated by its direct applicability for localisation of cancer metastases.
See Schoppe et al, Nature Communications
-

The Polymenidou group publishes their review on liquid-liquid phase separation in neurodegeneration
Phase Separation and Neuro-degenerative Diseases: A Disturbance in the Force
Many proteins found in pathological aggregations, the hallmark of neuro-degenerative diseases, have been shown to undergo reversible liquid-liquid phase separation under the right conditions. This suggests that aberrant phase separation may trigger the protein aggregation seen in neurodegeneration.
This review addresses similarities and differences among four proteins that are both known to undergo phase separation and are found as aggregations in neuro-degenerative diseases. In addition, future directions in this field are discussed that will contribute to elucidating the molecular mechanisms underlying aggregation and neurodegeneration.
See Zbinden, Pérez-Berlanga et al, Developmental Cell
-

The Polymenidou group receives competitive EU Consortium Grant
As coordinator of the ImageTDP43 consortium, the Polymenidou group has been awarded a competitive EU Consortium Grant from the 'EU Joint Programme - Neurodegenerative Disease Research (JPND).
The project 'ImageTDP43: Imaging heterogeneous TDP-43 neuropathologies' aims at developing TDP-43 PET ligands that will allow to accurately detect and monitor TDP-43-related neurodegenerative progression in diseases such as amyotrophic lateral sclerosis (ALS) and frontotemporal lobar degeneration with TDP-43 pathology (FTLD-TDP).The ImageTDP43 consortium brings together world-class academic partners from Fondazione Santa Lucia (Italy), Skåne University Hospital (Sweden) and Erasmus Medical Center (Netherlands), as well as industry partner AC Immune (Switzerland).
Please also read AC Immune's press release, which can be found here. -
Bernd Bodenmiller appointed as Dual Professor for Quantitative Biomedicine at UZH and ETH Zurich
We are excited to announce that Bernd Bodenmiller has been appointed as Dual Professor for Quantitative Biomedicine at the UZH and at ETH Zurich, starting from 1 October 2020. Please find the press release here:
https://ethz.ch/de/news-und-veranstaltungen/eth-news/news/2020/09/neue-professorinnen-und-professoren-ernannt.html.Bernd Bodenmiller’s dual affiliation is an important step towards the optimal positioning of DQBM within the Zurich quantitative biomedical community, in order to fulfill our mission to develop next-generation precision medicine in Zurich. We are excited about the new opportunities for scientific synergies between ETHZ and UZH resulting from this dual affiliation and look forward to the journey ahead.
-

The Kümmerli group shows the potential of combining antivirulence drugs with antibiotics against an opportunistic human pathogen
Combining antibiotics with antivirulence compounds can have synergistic effects and reverse selection for antibiotic resistance in Pseudomonas aeruginosa
This microbiology study reveals that compounds that disarm bacterial pathogens by targeting their virulence factors can be potent adjuvants to antibiotics, as they remain active against - and can reduce the selective advantage of - antibiotic resistant strains.
See Rezzoagli, Archetti et al., PLOS Biology
*Figure 4: Drug interaction heatmaps for antibiotic-antivirulence combination treatments.
-

The Bodenmiller group receives SNF support to study primary immune response & memory in COVID-19 patients
Together with Prof. Dr. med. Onur Boyman and Dr. med. Jakob Nilsson at the University Hospital Zurich andProf. Dr. Daniel Pinschewer at the University of Basel, the Bodenmiller group has received research project support from SNF NRP78 to contribute to our understanding of COVID-19. The project will focus on primary immune responses and the initial memory phase in mild vs. serious COVID-19 cases.
Please find the official announcement on the SNF website here.
-

The Bodenmiller group publishes their collaborative work on CD4+ T Helper Cell response heterogeneity
High-Dimensional T Helper Cell Profiling Reveals a Broad Diversity of Stably Committed Effector States and Uncovers Interlineage Relationships
This study resulted from a collaboration with the research group of Prof. Dr. Manfred Kopf at ETH Zurich.
CD4 + T helper (Th) are known to orchestrate immune responses. Based on the expression of signature cytokines and transcription factors, several Th subsets have previously been defined. In this study, CyTOF was used to systematically explore the diversity of Th cell responses generated both in vitro and in animal disease models, revealing a broad diversity in effector states with distinct cytokine footprints. -

Prof. Dr. Bjoern Menze and his team join the DQBM
The DQBM is delighted to announce that Prof. Dr. Bjoern Menze will join the DQBM Faculty as Full Professor for Biomedical Image Analysis and Machine Learning, funded by the Helmut Horten Foundation.
Prof. Menze studied physics in Heidelberg and Uppsala and received his PhD in Heidelberg in 2007. As postdoctoral researcher, he worked at renowned research institutions, including Harvard Medical School in Boston, MIT in Cambridge and ETH Zurich. In 2013, Prof. Menze was appointed a W2 professorship at the Technical University of Munich (TUM), where he has been researching and teaching since. In 2019, Prof. Menze was appointed W3 Professor for Image-Based Biomedical Modelling at the Munich School of BioEngineering and the Zentralinstitut fuer translationale Krebsforschung (TranslaTUM).We look forward to welcoming the Menze Group to our Department later this year!
---
Photo: © Andreas Heddergott / TU Muenchen -

The Kümmerli group shows how biotic and abiotic factors affect competitive dynamics between co-infecting human pathogens
Strain background, species frequency and environmental conditions are important in determining population dynamics and species co-existence between ;Pseudomonas aeruginosa and Staphylococcus aureus
Although bacterial communities in infections are typically diverse, little is known about how ecology affects inter-pathogen competition. Here, ecological theory was applied to understand how biotic and abiotic factors affect interaction patterns between two human pathogens that often co-occur in polymicrobial infections: Pseudomonas aeruginosa and Staphyloccocus aureus. This study revealed that ecological details such as strain background, species frequency and environmental conditions play important roles in the competitive dynamics between these co-infecting pathogens, suggesting that to truly understand polymicrobial infections, an integrative approach combining molecular and ecological aspects is essential.
*Figure 3 shows that the competitive ability of Pseudomonas aeruginosa depends on the Staphylococcus aureus strain genetic background. ;
See Niggli & Kümmerli, Applied & Environmental Microbiology
-
The Kümmerli group shows that competition overrides inter-species cooperation in bacteria
Antagonistic interactions subdue inter‐species green‐beard cooperation in bacteria
Cooperative behaviour among individuals is common, despite apparent fitness costs for the individual. The green‐beard mechanism can foster such cooperation: When a set of linked genes encodes both a cooperative trait and a phenotypic marker (green beard), carriers of that cooperative trait can selectively direct their cooperative actions to other carriers of this trait. This paper explored an extreme green‐beard scenario between two unrelated bacterial species P. aeruginosa and B. cenocepacia, which both produce the public good pyochelin (the cooperative trait) and an iron-pyochelin uptake receptor (the green beard). Competition experiments between these phylogenetically unrelated species revealed that the green‐beard cooperation effect collapses in this scenario, indicating that selection for competitive traits might overrule selection for cooperation in inter‐species interactions.
See Sathe & Kümmerli, Journal of Evolutionary Biology
-

The Krauthammer group publishes their work on deep learning-based multimodal fusion techniques to reduce annotation burden
Reducing Annotation Burden Through Multimodal Learning
This study examined deep learning-based multimodal fusion techniques for the combined classification of radiological images and associated text reports, comparing the classification performance of three prototypical multimodal fusion techniques. The experiments demonstrate the potential of multimodal fusion methods to yield competitive results using less training data than their unimodal counterparts. This suggests that the potential of multimodal learning decreases the need for labeled training data, which results in a lower annotation burden for domain experts.
See Lopez et al., Frontiers in Big Data
-

The Krauthammer Group publishes their work on machine learning models for assessing the quality of online health information.
AutoDiscern: rating the quality of online health information with hierarchical encoder attention-based neural networks
When facing health problems, patients increasingly turn to search engines and online information before or instead of talking to their doctor. However, as online health information is often of poor quality, this practice leads to potential misinformation. In this study, various machine learning models were built to evaluate the quality of online health information using the DISCERN criteria, which were developed at University of Oxford. The results suggest that automating the quality assessment of online health information is feasible, representing an important step towards enabling patients to become informed partners in the health process.
See Kinkead et al. BMC Medical Informatics and Decision Making
-

The Kümmerli group publishes their collaborative work on iron-driven phytopathogen control by natural rhizosphere microbiomes
Competition for iron drives phytopathogen control by natural rhizosphere microbiomes
This work results from a collaboration between the Kümmerli group and Alex Jousset, Zhong Wei, Shaohua Gu and others.
Soil-borne pathogenic bacteria are a global threat to food production, but due to the complexity of interactions between plants, their pathogens and the plant microbiome, plant infections are difficult to control. This study combined DNA-based soil microbiome analysis with in vitro and in planta bioassays to show that competition for iron via secreted siderophore molecules is a good predictor of microbe-pathogen interactions and plant protection. The results suggest that pathogen-suppressive microbiome members produce siderophores that the pathogen cannot use, establishing a causal mechanistic link between microbiome-level competition for iron and plant protection. This link provides opportunities to use siderophore-mediated interactions as a tool for microbiome engineering and pathogen control.
See Gu et al, Nature Microbiology
-

Magdalini Polymenidou awarded Franco Regli Prize 2018/2019
We congratulate Magdalini Polymenidou for being awarded the 2018/2019 Franco Regli Prize (2nd Prize ex æquo) for her work: TDP-43 extracted from frontotemporal lobar degeneration subject brains displays distinct aggregate assemblies and neurotoxic effects reflecting disease progression rates. Nature Neuroscience, Vol. 22, January 2019, 65–77.
Please find the announcement here -

The Krauthammer group publishes a data anonymization reference classification merging legal and technical considerations
Lost in Anonymization - A Data Anonymization Reference Classification Merging Legal and Technical Considerations
This work results from a collaboration between Michael Krauthammer, Daniel Stekhoven (Clinical Bioinformatics at the ETH Zurich core facility NEXUS Personalized Health Technologies) and Kerstin Vokinger (UZH).
Technological advances have made it possible to explore the wealth of health data collected in electronic health records (EHR) and other health-related data sources, with the aim of improving innovation and quality in medicine. These advances have led to the concern that the use of health data for publicly-funded research may expose patients personal information. This study i) analysed how different regulations implemented in the US, EU, and Switzerland to protect patient privacy distinguish between different levels of anonymization of health data, and ii) assessed whether and how these levels align with technical advancements.
See K.N. Vokinger et al., 2020, The Journal of Law, Medicine & Ethics -

Johanna Wagner-Albrecht (Bodenmiller group) wins the 2020 Faculty of Science's Annual Thesis Award
We congratulate Johanna Wagner-Albrecht from the Bodenmiller group for winning the 2020 Annual Thesis Award (Jahrespreis 2020 der Mathematisch-naturwissenschaftlichen Fakultät) for her dissertation: ''Single-Cell Proteomic Characterization of the Tumor and Immune Ecosystem of Human Breast Cancer with Focus on Metastatic Potential''.
-

Michael Krauthammer shares his vision on using AI to generate medical evidence from real world health data in UZH magazine
Is AI cleverer than us?
Today, Artificial Intelligence (AI) is widely used in the healthcare sector, relieving the burden on doctors and supporting them in taking medical decisions. Michael Krauthammer's vision for healthcare is to create a medical data repository that allows data to be exchanged worldwide, to supplement insights from clinical trials with data from daily medical practice. This would broaden doctors' horizons beyond their own patients to hundreds of thousands of patients worldwide. As the task of building up and maintaining this hypothetical medical data repository with anonymized patient data is highly complex, it will require the help of AI.
Please find the UZH News article here---
Image: Zentrale Informatik, Mels
-

The Bodenmiller group publishes their review on profiling cell signaling networks at single-cell resolution
Profiling cell signaling networks at single-cell resolution
Heterogeneity of signalling networks is at the basis of many biological processes, such as cell differentiation and drug resistance. Recent developments in multiplexed single-cell measurement technologies enabled evaluation of this heterogeneity. This review categorizes single-cell signaling network profiling approaches by their methodology, coverage, and application, and discusses the pros and cons of each technology. Furthermore, computational tools for network characterization using single-cell data are described. Finally, potential confounding factors that need to be considered in single-cell signaling network analyses are discussed.
See Lun & Bodenmiller, Molecular & Cellular Proteomics
-

The Bodenmiller group publishes their work linking imaging mass cytometry to breast cancer genomics
Imaging mass cytometry and multiplatform genomics define the phenogenomic landscape of breast cancer
A better understanding of how genomic alterations influence cell phenotypes and the structure of tumor ecosystems will likely allow the identification of biomarkers and the development of new treatments. In this study, Raza Ali, Hartland Jackson and colleagues coupled imaging mass cytometry (IMC) to multi-platform genomics to study how genomic alterations shape breast tumor ecosystems, improving our understanding of how genomic alterations affect tumor cell phenotypes.
-

The Kümmerli group publishes their review on harnessing bacterial interactions to manage infections
Harnessing bacterial interactions to manage infections: a review on the opportunistic pathogen Pseudomonas aeruginosa as a case example
During an infection, bacterial pathogens can interact in a variety of ways, from cooperative sharing of resources to hostile competition. This review focuses on the relevance of these social interactions during opportunistic infections using the human pathogen P. aeruginosa as an example and shows how a deeper understanding of bacterial social dynamics can inspire new treatment approaches and improved, more sustainable infection management strategies.
See Rezzoagli et al., Journal of Medical Microbiology
-

The Bodenmiller group publishes their work on the single-cell pathology landscape of breast cancer
The single-cell pathology landscape of breast cancer
See Jackson, Fischer et al, NatureLink to UZH's news release
-

The Bodenmiller group, in collaboration with the Krishnaswamy group, publishes a method to uncover axes of variation among single-cell cancer specimens
Uncovering axes of variation among single-cell cancer specimens
This work results from a collaboration between the Bodenmiller group and the Krishnaswamy group at Yale School of Medicine.
William S. Chen and Nevena Zivanovic contributed equally to this work.
This work was co-supervised by Bernd Bodenmiller and Smita Krishnaswamy.
See Chen, Zivanovic et al, Nature Methods -

DQBM's roundup of 2019
DQBM was founded on 1.1.2019. Our joint mission is to foster research and education at the interface of biomedical research, biotechnology, and computational biology, to develop the foundations of next generation precision medicine. Ultimately, our goal is to advance precision medicine for the benefit of patients.
-

Bernd Bodenmiller awarded ERC Consolidator Grant
We congratulate Prof. Dr. Bernd Bodenmiller, who has been awarded a prestigious ERC Consolidator Grant worth 2M EUR for his project 'Precision Motifs': Analysis of functional tissue motifs for precision medicine in metastatic breast cancer.
-
Julien Weber (Polymenidou group) wins Best Lab Manager Award 2019
We congratulate Julien Weber from the Polymenidou group, who was voted Best Lab Manager 2019!
Please find the announcement here -
The Kümmerli group shows that homogeneous rather than specialised bacterial behavioral repertoires prevail in natural Pseudomonas communities
Positive linkage between bacterial social traits reveals that homogeneous rather than specialised behavioral repertoires prevail in natural Pseudomonas communities
See Kramer et al, FEMS Microbiology Ecology
-
The Kümmerli group publishes their review on bacterial siderophores in community and host interactions
See Kramer et al, Nature Reviews Microbiology
Link to paper: https://www.nature.com/articles/s41579-019-0284-4
-
Bernd Bodenmiller selected for the Analytical Scientist's 2019 Power List Top 100
We congratulate Prof. Dr. Bernd Bodenmiller, who was selected for the Analytical Scientist's Power List 2019 Top 100.
Please find the Top 100 here:
https://theanalyticalscientist.com/powerlist/2019 -
The Kümmerli group publishes their work on genetic constraints of siderophore cooperation exploitation in Burkholderia cenocepacia
See Sathe et al, Evolution Letters
Link to paper: https://doi.org/10.1002/evl3.144
-
Magdalini Polymenidou appointed Associate Professor for Biomedicine, in particular Molecular Pathogenesis of Neurodegeneration
We congratulate Prof. Dr. Magdalini Polymenidou on her appointment as Associate Professor for Biomedicine, in particular Molecular Pathogenesis of Neurodegeneration, effective October 1, 2019.
Please see the official announcement here (in German only):
https://www.news.uzh.ch/de/articles/2019/ernennungen-september-2019.html -
The Krauthammer group publishes their work on neural network-based models versus logistic regression for predicting readmission
See Allam et al, Scientific Reports
Pubmed link to paper: https://www.ncbi.nlm.nih.gov/pubmed/31243311
-
The Bodenmiller group publishes a human kinome- and phosphatome-wide screen revealing overexpression-induced effects on cancer-related signaling
See Lun et al, Molecular Cell
Pubmed link to paper: https://www.ncbi.nlm.nih.gov/pubmed/31101498
-
The Kümmerli group publishes their work on the impact of Pseudomonas aeruginosa social interactions on host colonization
See Rezzoagli et al, The ISME Journal
Pubmed link to paper: https://www.ncbi.nlm.nih.gov/pubmed/31123320
-
The Bodenmiller group finds that breast cancer ecosystems are linked to poor prognosis and immunosuppression.
See Wagner et al, Cell,
www.cell.com/cell/fulltext/S0092-8674(19)30267-3
-
The Polymenidou group publishes their work on RNA binding of FUS
The Solution Structure of FUS Bound to RNA Reveals a Bipartite Mode of RNA Recognition with Both Sequence and Shape Specificity
This work resulted from a collaboration with the Allain group at ETH Zurich.
See Loughlin et al, Molecular Cell
-
The Polymenidou group publishes their work on TDP-43 strains
TDP-43 extracted from frontotemporal lobar degeneration subject brains displays distinct aggregate assemblies and neurotoxic effects reflecting disease progression rates
See Laferriere et al, Nature Neuroscience
Please also see this accompanied news and views article
and the commentary on Alzforum.
-

Bernd Bodenmiller receives Friedrich Miescher Award
Bernd Bodenmiller has been selected to receive the prestigious Friedrich Miescher Award 2019 for his research at the Institute of Molecular Life Sciences of the University of Zurich. The prize is Switzerland's highest distinction for young scientists performing outstanding research in the field of biochemistry.
Please find the press release here.
-
The Department of Quantitative Biomedicine has been founded!
We are a new department that will foster research and education at the interface of biomedical research, biotechnology, and computational biology to develop the foundations of next generation precision medicine. Our groups currently cover the topics of cancer, neurodegenerative diseases and infectious diseases. Enjoy browsing our website to learn more about our work.
